MENU
DK | DKK
DK | DKK
No results found
Search Suggestions

Efficient & Automated qPCR Setup Without Cross Contamination
Lab Academy
- Molecular Biology
- Liquid Handling Workstations
- Amplification & PCR
- Reproducibility
- Contamination
- Automation
- Essay
Standard PCR combined with a fluorescent reporter monitoring, the real-time quantitative PCR (qPCR) allows to amplify, detect, and quantify the targeted molecule in a single step. However, manual qPCR set-up is time-consuming, tedious, and requires a lot of practice, because the most probable source of imprecision is the operator itself. Learn how automation will increase your efficiency.
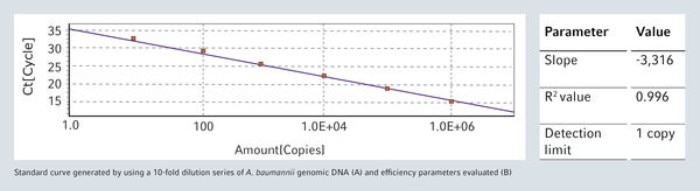
Automated Liquid Handling systems are ideal for reliable and reproducible set-up of sensitive real-time quantitative PCR (qPCR) assays in daily routine, without the risk of cross contamination. As example, a very sensitive qPCR assay developed for Acinetobacter baumannii has been selected and set-up with epMotion workstations. Performances of this assay are demonstrated by generating a standard concentration curve of genomic DNA (gDNA).
Read more
Read less
Besides issues with reproducibility, qPCR reactions can also be affected by nucleic acid contamination, leading to false positive results.
The three possible sources of contamination are:
Precautions can be taken to reduce the risk of contamination: uracil DNA glycosylase (UDG) can be used to prevent DNA carryover contamination between reactions, materials can be systematically decontaminated, and separate workstations for each step of the qPCR process can be designated to create an efficient workflow. However, those measures do not eliminate the major cause of false positive results: the accidental contamination with positive samples during Liquid Handling.
The three possible sources of contamination are:
- cross-contamination between samples
- contamination from laboratory equipment and
- carryover contamination by amplified products from previous qPCRs.
Precautions can be taken to reduce the risk of contamination: uracil DNA glycosylase (UDG) can be used to prevent DNA carryover contamination between reactions, materials can be systematically decontaminated, and separate workstations for each step of the qPCR process can be designated to create an efficient workflow. However, those measures do not eliminate the major cause of false positive results: the accidental contamination with positive samples during Liquid Handling.
Read more
Read less
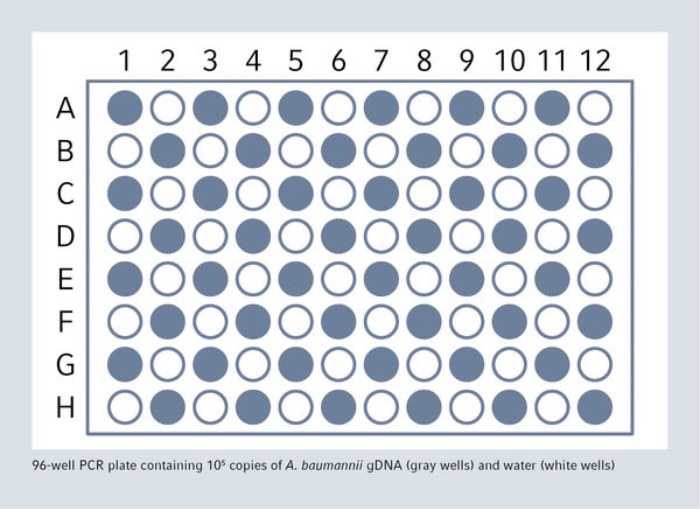
In order to demonstrate that cross-contamination will not occur, an FDA guideline proposes to use high positive samples in series alternating with negative samples. The epMotion was used for a complete qPCR set-up without cross-contamination, 48 high positive samples (containing 105 copies of A. baumanii gDNA) and 48 negative samples (containing water) have been dispensed in the 96-well PCR plate in a chessboard pattern.
Read more
Read less
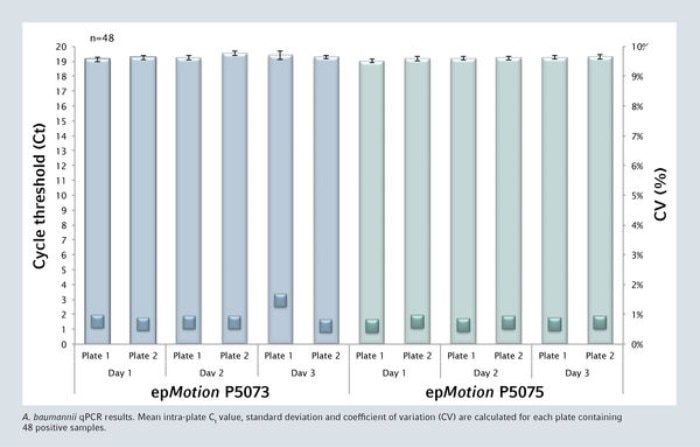
All negative samples were negative and the complete process is highly reproducible. The mean intra-plate Ct value for the 48 positive samples is very consistent with a coefficient of variation never exceeding 1.5%.
Click here to read the whole Applicaiton Note.
Click here to read the whole Applicaiton Note.
Read more
Read less

