MENU
MY | MYR
Logged in as:
MY | MYR
No results found
Search Suggestions
UV-Vis Spectrophotometry – Easy and Quick Quantification of Nucleic Acids
Natascha Weiß Lab Academy
- Cell Biology
- Photometers
- Photometry
- Essay
The absorbance measurement of a nucleic acid solution at only four wavelengths is sufficient to determine its concentration and to obtain an insight into its purity.
Nucleic acids are isolated from sample material such as cells and subsequently employed in further laboratory experiments. For this purpose, it is advisable to determine the concentration of the purified DNA or RNA as well as verify their purity. In this way, accurately defined amounts of the nucleic acid will enter the downstream application, and any contaminations that may impact a sensitive reaction or assay are detected in real time.
UV-Vis spectrophotometry is an easy, quick and time-tested method to achieve these objectives. In the range of 260 nm, nucleic acids show a characteristic absorbance peak (figure 1). This absorbance value is therefore used to calculate nucleic acid concentrations. According to the Lambert-Beer law, two parameters are required for the calculation of the concentration of a sample: the optical path length (= light path length (L)) and the molar extinction coefficient (material and wavelength-specific constant) of the sample to be measured. The specific factor (F) can be calculated when using a standard cuvette with a light path of 1 cm. This factor is then multiplied by the measured absorbance value (A) in order to arrive at the concentration (C) of the sample solution. Molar extinction coefficients and specific factors, respectively, are typically available in the literature. The factor for dsDNA, for example, is 50 µg/mL (RNA: 40 µg/mL), and it is by definition equivalent to one absorbance unit. these data.
UV-Vis spectrophotometry is an easy, quick and time-tested method to achieve these objectives. In the range of 260 nm, nucleic acids show a characteristic absorbance peak (figure 1). This absorbance value is therefore used to calculate nucleic acid concentrations. According to the Lambert-Beer law, two parameters are required for the calculation of the concentration of a sample: the optical path length (= light path length (L)) and the molar extinction coefficient (material and wavelength-specific constant) of the sample to be measured. The specific factor (F) can be calculated when using a standard cuvette with a light path of 1 cm. This factor is then multiplied by the measured absorbance value (A) in order to arrive at the concentration (C) of the sample solution. Molar extinction coefficients and specific factors, respectively, are typically available in the literature. The factor for dsDNA, for example, is 50 µg/mL (RNA: 40 µg/mL), and it is by definition equivalent to one absorbance unit. these data.
Read more
Read less
Lambert-Beer law
| C = A/(L x Ɛ ) | C = Concentration |
| F = 1/Ɛ (for a light path of 1 cm) | A = Absorbance |
| L = Optical (light) path length | |
| - C = A x F | Ɛ = Molar extinction coefficient(sample and wavelength-specific) |
| F = Factor |
Read more
Read less
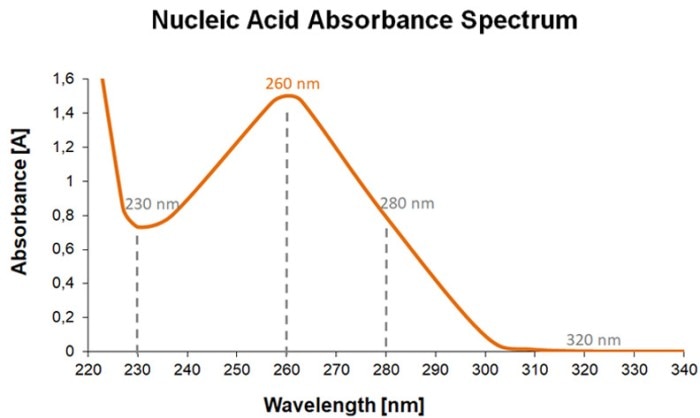
Figure 1: Absorbance spectrum of nucleic acids with relevant wavelengths and an example describing the calculation of the concentration of a dsDNA sample.
In order to be able to make a statement about the purity of a nucleic acid sample, the absorbance at wavelengths other than 260 nm needs to be determined (figure 1). The quotients derived from the absorbance values measured at 260 nm, 280 nm and 230 nm (A260/A280 and A260/A230) constitute the purity ratios which help identify possible contaminations. Impurities such as proteins and traces of reagents that were used during the purification process yield a different absorbance spectrum from that of nucleic acids and thus influence the purity ratios (figure 2). The A260/A280 ratio of pure nucleic acid solutions will be approximately 1.8 - 2.0, whereas the ratio A260/A230 will typically show values in the range of 2.0 – 2.5.
Read more
Read less
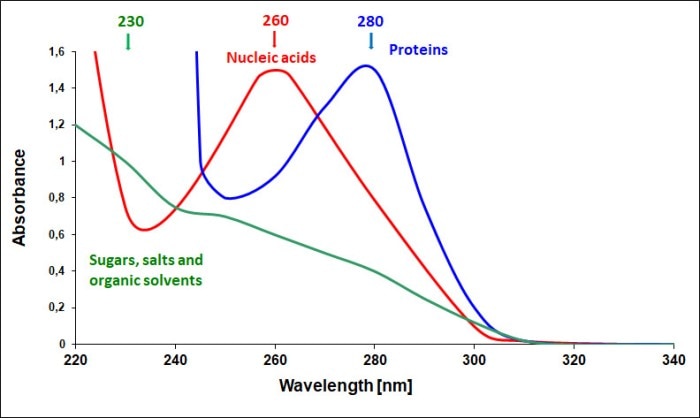
Figure 2: Absorbance spectra of nucleic acids and possible contaminants
A fourth wavelength is used to determine the background. It is measured at or above 320 nm as neither nucleic acids nor organic contaminants absorb light at this wavelength. Absorbance measured in this range may indicate the presence of particles or air bubbles within the sample. Even a smudged cuvette is capable of eliciting a background reading. Modern photometers offer the option of activating a background correction feature which will effect automatic subtraction of any background absorbance from all other measured values.
In addition, the complete spectrum of a nucleic acid sample may be captured (typically between 220 and 320 nm). Comparisons with the spectrum obtained from a pure nucleic acid solution will enable the detection of possible errors during the measurement process as well as the presence of contaminants within the sample.
In addition, the complete spectrum of a nucleic acid sample may be captured (typically between 220 and 320 nm). Comparisons with the spectrum obtained from a pure nucleic acid solution will enable the detection of possible errors during the measurement process as well as the presence of contaminants within the sample.
Read more
Read less
Read more
Read less

