-
-
-
-
- Forum Labo 2025
- Advanced Therapies Week (ATW) 2025
- SLAS Europe 2025
- Bioprocessing Summit Europe 2025
- Medlab Middle East 2025
- SLAS International 2025
- Biologics World Nordics 2025
- ASIA LABEX: The Lab Show 2025
- BioProcess International Europe 2025
- ISEV 2025
- Future Labs Live 2025
- DataHow Symposium 2025
- Cell 2025
- LabDays 2025
- ILMAC 2025
- ASIA LABEX: The Lab Show 2025
-
-
-
-
- Forum Labo 2025
- Advanced Therapies Week (ATW) 2025
- SLAS Europe 2025
- Bioprocessing Summit Europe 2025
- Medlab Middle East 2025
- SLAS International 2025
- Biologics World Nordics 2025
- ASIA LABEX: The Lab Show 2025
- BioProcess International Europe 2025
- ISEV 2025
- Future Labs Live 2025
- DataHow Symposium 2025
- Cell 2025
- LabDays 2025
- ILMAC 2025
- ASIA LABEX: The Lab Show 2025
-
- Benchtop Centrifuges
- Floor-Standing Centrifuges
- Refrigerated Centrifuges
- Microcentrifuges
- Multipurpose Centrifuges
- High-Speed Centrifuges
- Ultracentrifuges
- Concentrator
- IVD Products
- High-Speed and Ultracentrifuge Consumables
- Centrifuge Tubes
- Centrifuge Plates
- Device Management Software
- Sample and Information Management
-
- All Pipettes, Dispensers & Automated Liquid Handlers
- Mechanical Pipettes
- Electronic Pipettes
- Multi-Channel Pipettes
- Positive Displacement Pipettes & Dispensers
- Automated Pipetting
- Bottle-Top Dispensers
- Pipette Controllers
- Pipette Tips
- Automation Consumables
- Dispenser & Pipette Accessories
- Automation Accessories
- Dispenser & Pipette Services
You are about to leave this site.
Please be aware that your current cart is not saved yet and cannot be restored on the new site nor when you come back. If you want to save your cart please login in into your account.
Loop-Mediated Isothermal Amplification (LAMP) of Human Apolipoprotein L1 Gene
Nina Egeler, Steffen Riethmüller, Arora Phang Lab Academy
- Molecular Biology
- Amplification & PCR
- Mixers & Shakers
- PCR Cyclers
- BioNews article
The recent COVID-19 pandemic showed a silver lining in the molecular biology world by spurring on improvements in many methods especially in the detection of nucleic acids. “Loop-mediated isothermal amplification” (LAMP) has become a popular detection method due to its operation simplicity and rapidity. Using the WarmStart® Colorimetric LAMP 2X Master Mix (New England Biolabs®), the human apolipoprotein L1 gene (ApoL1) was detected in just 30 min with the Eppendorf ThermoMixer® C and the Eppendorf ThermoTop®. Additionally, even very low template DNA amounts of 0.1 ng were detectable by simply extending the incubation time to 45 min.
The amplification of nucleic acids is a widespread tool for the detection of various organisms and pathogens. Besides commonly used PCR-based methods, isothermal amplification offers an alternative for the detection of nucleic acids. Recently, the interest in isothermal amplification grew rapidly because of the COVID-19 pandemic. Some isothermal amplification methods are now already in use as PoC-NAT-tests (Point of care nucleic acid amplification tests) for the Coronavirus.
One of these methods is loop-mediated isothermal amplification (LAMP). LAMP was developed in 2000 by Notomi et al. [1]. During the reaction, a dumbbell-shaped DNA loop is created through primer annealing and the amplification is performed by polymerases with strand-displacement activity. This eliminates the need for an additional denaturation step and leads to continuous amplification of DNA under isothermal conditions. By adding reverse transcriptase to the assay, LAMP can also be used for the detection of RNA.
Successful LAMP reactions can be instantly observed through turbidimetry or colorimetric dyes without the necessity of an additional time-consuming step like gel electrophoresis. In this Application Note, we show the detection of the human ApoL1 gene with two different polymerases and the following examination of its detection sensitivity.
Materials and methods
Parts of ApoL1 were amplified in 0.1 mL Eppendorf PCR Tube Strips from human genomic DNA (Promega® ). See full Application Note 454 for detailed information*.
Assessment of test sensitivity
To observe how quickly lower template DNA amounts can be detected, the template DNA was diluted, and the color of the colorimetric dye was examined at different timepoints. The final samples contained the WarmStart Colorimetric LAMP 2X Master Mix with 0.001 ng, 0.01 ng, 0.1 ng, 1 ng, 10 ng, 50 ng, or 100 ng of template DNA. The samples were incubated at 65 °C on the Eppendorf ThermoMixer C with the ThermoTop for up to 60 min.
Assessment of LAMP technique
To assess the viability of the LAMP technique, a second experiment was performed using a different reaction kit (Isothermal Master Mix, OptiGene) without a colorimetric dye. The LAMP reactions containing the master mix and 50 ng of template DNA were incubated at 65 °C on the Eppendorf ThermoMixer C with the ThermoTop for 45 min. Additionally, a PCR was performed using the Mastercycler® X50a (Eppendorf) for comparison.
Results and discussion
Assessment of test sensitivity
The colorimetric dye contained in the WarmStart Colorimetric LAMP 2X Master Mix allowed instant identification of amplified DNA during the incubation of the LAMP reaction. This in turn, allowed qualitative assessment of the sensitivity of the LAMP assay. As expected, the reaction took longer to change color with less template DNA (Fig. 1).
Read more
Read less
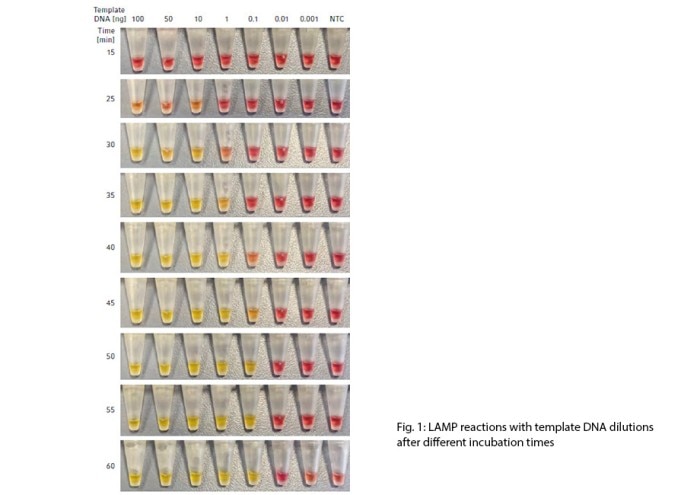
Under tested conditions, this protocol showed a sensitivity limit of 0.1 ng template DNA, which turned positive yellow after 45 min. The time assessment range was set at 60 min termination when the color of the no template control (NTC) was observed to have started to change, indicating that the assay was no longer reliable with further incubation.
Assessment of LAMP technique
To assess the viability of the LAMP technique for DNA detection, the ApoL1 gene amplification was repeated using another reaction kit without a colorimetric dye. The results, processing times, and costs were compared to the PCR technique to analyze respective pros and cons.
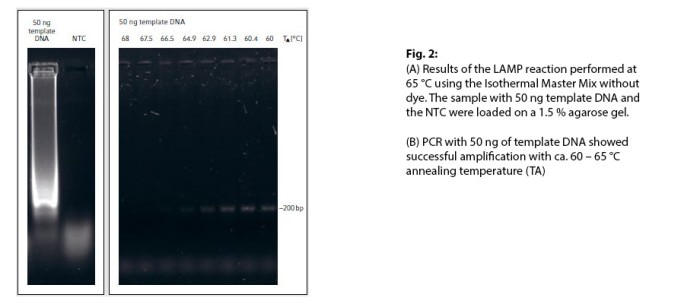
Detection of the ApoL1 gene was successfully achieved using the Isothermal Master Mix, as was shown by the characteristic smear on the gel (Fig. 2A).
The reaction took 45 min. In comparison, the PCR technique used to achieve a positive DNA amplification of the ~200 bp amplicon (Fig. 2B) took 105 min. Using fast PCR reagents and an optimized PCR protocol can further shorten the total runtime. However, an optimized LAMP kit pre-mixed with an inexpensive colorimetric dye (e.g. hydroxynaphthol blue) will be probably cheaper and definitely faster since it does not require additional equipment and saves the step of running a gel.
Read more
Read less
The LAMP technique proves to be a viable method for DNA detection, as shown herein whereby the ApoL1 gene was positively identified after 30 min. Moreover, this technique has high sensitivity of up to 0.1 ng template DNA and fast reaction completion time. Furthermore, end-point visualization of LAMP reactions can be carried out via simple colorimetric detection. This makes the additional step of running an agarose gel unnecessary, saving users the extra step and time compared to end-point PCR. In summary, the LAMP technique is a quick and simple alternative for the detection of nucleic acids that can be performed using a thermal cycler as well as an Eppendorf ThermoMixer C.
Literature
[1] Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N & Hase T (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Research, 28 (12).
Note: Eppendorf SE reserves the right to modify its products and services at any time. This Application Note is subject to change without notice. Although prepared to ensure accuracy, Eppendorf SE assumes no liability for errors, or for any damages resulting from the application or use of this information. Viewing Application Notes alone cannot as such provide for or replace reading and respecting the current version of the operating manual.
Read more
Read less
