MENÚ
ES | EUR
ES | EUR
-
- Centrífugas de sobremesa
- Centrífugas de suelo
- Centrífugas refrigeradas
- Microcentrífugas
- Centrífugas multiuso
- Centrífugas de alta velocidad
- Ultracentrífugas
- Concentrador
- Productos IVD
- High-Speed and Ultracentrifuge Consumables
- Tubos de centrífuga
- Placas de centrífuga
- Gestión de dispositivos
- Gestión de muestras e información
-
- Pipeteo manual & dispensación
- Pipetas mecánicas
- Pipetas electrónicas
- Pipetas multicanal
- Pipetas de desplazamiento positivo y dispensadores
- Pipeteo automatizado
- Dispensadores de botella
- Controladores de pipeta
- Puntas de pipeta
- Consumibles de automatización
- Accesorios para dispensadores y pipetas
- Accesorios de automatización
- Servicios para dispensadores y pipetas
Sorry, we couldn't find anything on our website containing your search term.

How to select an E. coli strain
Academia de laboratorio
- Farmacia
- Biología molecular
- Bioprocesamiento
- Biotecnología
- Alimentos y bebidas
- Microbiología
- Salud y medicina
- Eficiencia
- Cultivo de microorganismos
- Amplificación y PCR
- Consumibles para cultivo celular
- Ensayo
This article provides insight into a range of genetically altered E. coli strains to help determine which strain is most suitable for your desired outcome or experiment.
Who hasn’t heard of Escherichia coli? E. coli is the celebrity of the microbe and scientific world, as it has enabled us to study and understand genetics and proteins like never before1,2,3 . Due to its fast high-density cultivation, ease of genetic manipulation and relatively simple scale-up procedure, E. coli plays a crucial role in molecular biology and in the industrial production of recombinant proteins and plasmid DNA4 . This is usually the first step for a lot of laboratory work including the research of gene function or protein interactions, drug toxicity studies, or the generation of vast quantities of proteins like insulin.
The E. coli strains used in the laboratory are genetically modified to further enhance their ability to aid gene cloning and protein expression. For example, E. coli strains that are easier to screen, have enhanced production or quality of recombinant proteins, or increased plasmid stability are available. As different strains are optimised for particular applications, selecting the best E. coli strain for a particular application can be challenging.
This article provides insight into a range of genetically altered E. coli strains to help determine which strain is most suitable for your desired outcome or experiment.
The E. coli strains used in the laboratory are genetically modified to further enhance their ability to aid gene cloning and protein expression. For example, E. coli strains that are easier to screen, have enhanced production or quality of recombinant proteins, or increased plasmid stability are available. As different strains are optimised for particular applications, selecting the best E. coli strain for a particular application can be challenging.
This article provides insight into a range of genetically altered E. coli strains to help determine which strain is most suitable for your desired outcome or experiment.
Leer más
Leer menos
Why do you need a specific E. coli strain?
Let’s first talk about plasmids. These genetic molecules are typically small circular DNA strands in the cytoplasm of bacteria or protozoa that can replicate independently from chromosomal DNA. Plasmids represent the basis for the majority of molecular biology applications and are routinely employed in the laboratory to modify or introduce genes into bacteria.
Genes of interest can be studied by isolating and inserting them into plasmids which are then transformed into bacterial hosts. The bacteria will then replicate and propagate this gene so that there are numerous copies. Cloning, genetic engineering, and the creation of recombinant proteins are all made possible by this technique. Ground-breaking research like the Human Genome Project and the creation of insulin - the first genetically engineered human drug - were all made possible thanks to these techniques5,6 .
E. coli is considered the standard bacterial host for molecular genetics as it has a high transformation efficiency, rapid growth and the ability to express proteins at high levels7 . However, there are now hundreds of different strains of E. coli to choose from, each with individual traits that are optimized for certain applications.
It’s therefore important that an appropriate strain is chosen to host your particular plasmid. A short search of the literature will turn up dozens of potential candidates for an appropriate E. coli strain to use as a host, with various advantages and disadvantages for all of them.
Let’s first talk about plasmids. These genetic molecules are typically small circular DNA strands in the cytoplasm of bacteria or protozoa that can replicate independently from chromosomal DNA. Plasmids represent the basis for the majority of molecular biology applications and are routinely employed in the laboratory to modify or introduce genes into bacteria.
Genes of interest can be studied by isolating and inserting them into plasmids which are then transformed into bacterial hosts. The bacteria will then replicate and propagate this gene so that there are numerous copies. Cloning, genetic engineering, and the creation of recombinant proteins are all made possible by this technique. Ground-breaking research like the Human Genome Project and the creation of insulin - the first genetically engineered human drug - were all made possible thanks to these techniques5,6 .
E. coli is considered the standard bacterial host for molecular genetics as it has a high transformation efficiency, rapid growth and the ability to express proteins at high levels7 . However, there are now hundreds of different strains of E. coli to choose from, each with individual traits that are optimized for certain applications.
It’s therefore important that an appropriate strain is chosen to host your particular plasmid. A short search of the literature will turn up dozens of potential candidates for an appropriate E. coli strain to use as a host, with various advantages and disadvantages for all of them.
Leer más
Leer menos
Which E. coli strain is optimized for gene cloning?
In 1973, Herbert Boyer and Stanley Cohen showed for the first time that two short strands of bacterial DNA could be linked together and transferred back to E. coli8 . Following from this, the era of molecular cloning began, opening up many new avenues of scientific research.
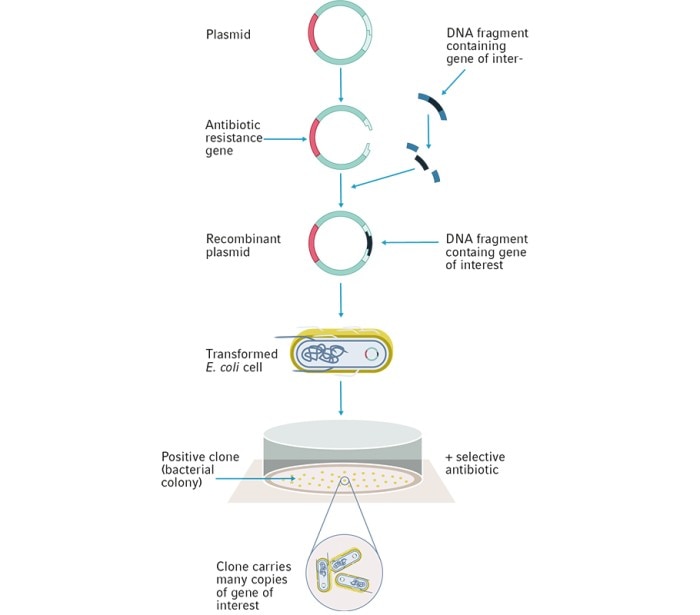
Today, E. coli continues to be used as the standard host for the storage and propagation of plasmids containing DNA sequences of interest (Figure 1). Increasingly optimised strains of E. coli have since been produced due to genetic engineering. The majority of these different E. coli strains are descendants of the K-12 and B strain isolates dating back from the 1920s9,10 , with mutations that confer a variety of properties to make them better suited for different requirements and applications.
In 1973, Herbert Boyer and Stanley Cohen showed for the first time that two short strands of bacterial DNA could be linked together and transferred back to E. coli8 . Following from this, the era of molecular cloning began, opening up many new avenues of scientific research.
Today, E. coli continues to be used as the standard host for the storage and propagation of plasmids containing DNA sequences of interest (Figure 1). Increasingly optimised strains of E. coli have since been produced due to genetic engineering. The majority of these different E. coli strains are descendants of the K-12 and B strain isolates dating back from the 1920s9,10 , with mutations that confer a variety of properties to make them better suited for different requirements and applications.
Leer más
Leer menos
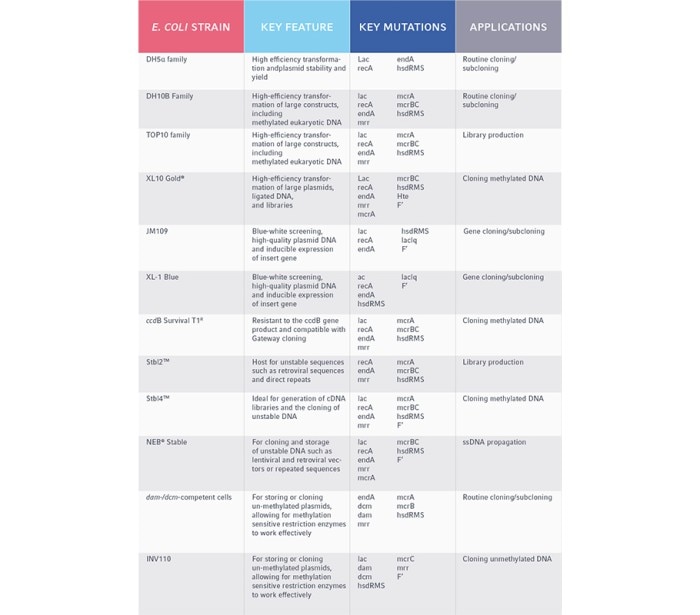
When deciding which E. coli strain may be suitable for you, it’s important to consider your methodology. For a typical gene cloning experiment, you may need a high rate of transformation efficiency or the capacity for a large plasmid for long genetic sequences. For a large sequencing experiment, you may require an E. coli strain that can handle hundreds of thousands of DNA base pairs in the form of bacterial artificial chromosomes (BAC), in order to create DNA libraries.
Common mutations and their associated properties:
Common mutations and their associated properties:
- lac: This mutation allows for blue/white screening to rapidly detect correctly transformed bacteria. Cells transformed with vectors containing the cloned DNA result in white colonies; while cells transformed with non-recombinant plasmids (i.e., only the vector) grow into blue colonies.
- recA or endA: These mutations disrupt the recombination system in E. coli, resulting in reduced recombination of plasmids into the bacterial genome and thus, increased plasmid stability. This helps to increase the chances that the plasmid is inherited as cells divide and replicate to ensure your DNA is propagated.
- Hte: This phenotype has an extremely high transformation efficiency. This is particularly useful if you have large plasmid constructs (>15kb).
- deoR: This mutation allows the cell to maintain large genetic constructs and improves the chance of cloning full-length cDNA sequences.
- F′: This mutation allows for the propagation of single-stranded DNA (ssDNA).
- dcm/dam: This mutation disrupts the DNA methyltransferase pathway. Strains with these mutations are useful for preparing unmethylated DNA, which may be important when trying to cut with methylation-sensitive restriction enzymes.
- hsdRMS: This mutation enables propagation of unmethylated non–E. coli DNA by protecting sequences like PCR amplicons from degradation.
- mcrA, mcrBC, and mrr: Protects methylated eukaryotic DNA from degradation, allowing for the uptake and propagation of methylated genes of interest.
- ccdB resistance: Strains with this mutation can be transformed with plasmids containing the toxic marker gene ccdB. This is useful for recombinant techniques that don’t use restriction enzymes.
Leer más
Leer menos
Table 1. Common E. coli strains and their gene cloning applications
Leer más
Leer menos
Which E. coli strains are optimized for recombinant protein expression?
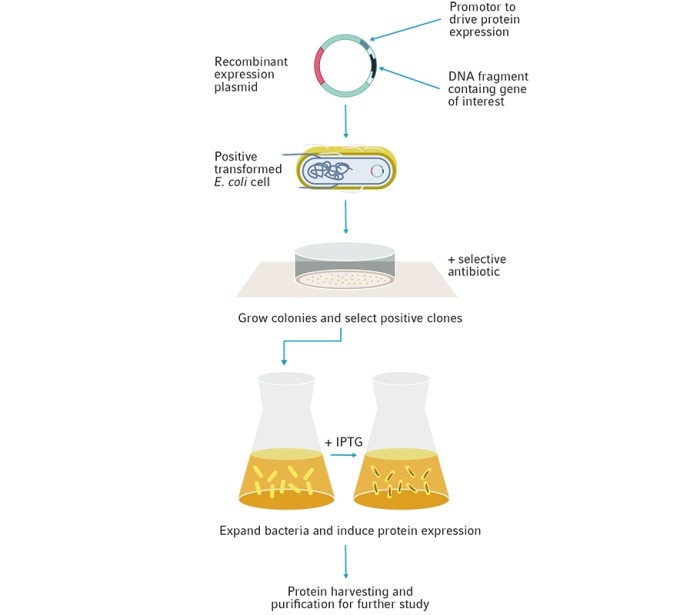
Production of recombinant proteins starts at the genetic level, where the protein's coding sequence is first extracted and cloned into an expression plasmid vector. This protein is then expressed and can be used for a variety of purposes, ranging from large-scale synthesis of biotherapeutic drugs to the study of gene regulation, protein structure, function and protein-protein interactions (Figure 2).
Production of recombinant proteins starts at the genetic level, where the protein's coding sequence is first extracted and cloned into an expression plasmid vector. This protein is then expressed and can be used for a variety of purposes, ranging from large-scale synthesis of biotherapeutic drugs to the study of gene regulation, protein structure, function and protein-protein interactions (Figure 2).
Leer más
Leer menos
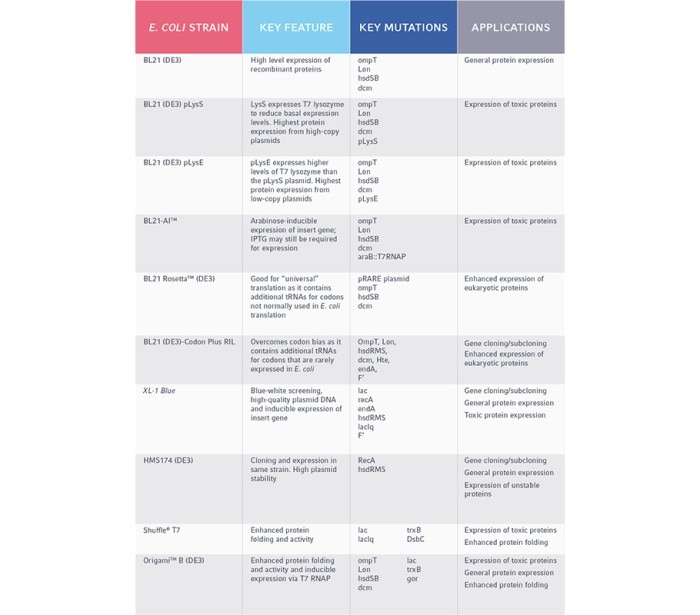
Due to its quick and effective growth, E. coli is one of the most widely used hosts for the overexpression of recombinant proteins. However, many challenges can arise when over-expressing a foreign protein in E. coli. High copy number plasmids with strong promoters will express proteins at much higher levels than the native host proteins, depleting the cell's resources and limiting growth. Additionally, some protein products may be toxic to the host when expressed, particularly those that are insoluble, act on DNA, or are enzymatically active. As a result, inducible promoter systems are frequently used to express recombinant proteins, enabling cells to be grown to the proper density before the recombinant protein is expressed. Furthermore, specialist E. coli strains can have mutations to assist with protein folding, transcriptional regulation, and improved production of eukaryotic proteins.
Since the expression of recombinant proteins still relies on the transformation of a plasmid, the mutations listed above still apply to E. coli selection for protein expression. However, there are also additional, specific mutations to encourage high protein levels.
Since the expression of recombinant proteins still relies on the transformation of a plasmid, the mutations listed above still apply to E. coli selection for protein expression. However, there are also additional, specific mutations to encourage high protein levels.
Leer más
Leer menos
Common mutations in E. coli strains for protein expression:
- DE3: Represents the T7 promoter, which drives higher expression of host cell’s endogenous RNA polymerases to result in more recombinant proteins. IPTG is used to induce this promoter, allowing for cells to be grown to an appropriate density before the recombinant protein is expressed.
- Lon: Strains deficient in the Lon protease have reduced proteolysis of the expressed proteins.
- OmpT: Deficient in the outer membrane protease OmpT, resulting in reduced proteolysis.
- hsdSB: In wild-type E. coli, DNA that does not contain methylation of certain sequences is recognized as foreign and degraded. This mutation disrupts DNA methylation and degradation, preventing the loss of your protein-expressing plasmid.
- gor or trxB: Strains have enhanced capacity to correctly fold proteins with multiple disulfide bonds in the cytoplasm.
- DsbC: The DsbC isomerase is an effective chaperone that can assist in the folding of target proteins, independent of disulfide bond formation.
- pRARE: This plasmid enhances the ability of the E. coli strain to express large quantities of eukaryotic proteins as it contains additional tRNAs for rare codons not normally used in E. coli translation.
- Rne131: Strains with this mutation lack a functional RNaseE and thus, have reduced capacity for mRNA degradation. This results in a longer transcript half-life for inserted DNA sequences and thus, higher protein production.
- pLys plasmid: Expresses T7 lysozyme which inhibits T7 RNA polymerase activity. This is to ensure that no recombinant protein is expressed until the promoter is induced by IPTG. This is particularly important if the inserted gene codes for a toxic protein.
- laclq: Similar to the pLys plasmid, this mutation ensures that inserted genes are not expressed unless specifically induced. This is accomplished by overproducing the lac repressor, which prevents the T7 RNA polymerase from being expressed.
Leer más
Leer menos
Table 2. Common E. coli strains and their applications in protein expression
Leer más
Leer menos
Summary
Gene cloning is the cornerstone of molecular biology. It is the first stage in producing vast amounts of any DNA sequence, which can later be employed in laboratories or for commercial applications. The importance of both gene cloning and recombinant protein production has increased rapidly across life science research, and in the development of diagnostic reagents and therapeutic drugs, as well as in food production and agriculture.
Advancements in the field of biotechnology have increased and facilitated a more efficient and productive system for gene cloning and the production of recombinant proteins – as demonstrated by the enormous diversity of E. coli strains that are now readily available. Enhanced protein folding, greater transcriptional control and improved plasmid stability have enabled efficient gene cloning and high-throughput protein expression for structural and functional studies, in addition to industrial production.
Whether you are producing cDNA libraries, cloning methylated DNA or producing toxic proteins, there is an E. coli strain to suit your purposes. The information in this guide should help you to select the ideal strain for your experimental requirements based on their mutations and the key features they enable.
Gene cloning is the cornerstone of molecular biology. It is the first stage in producing vast amounts of any DNA sequence, which can later be employed in laboratories or for commercial applications. The importance of both gene cloning and recombinant protein production has increased rapidly across life science research, and in the development of diagnostic reagents and therapeutic drugs, as well as in food production and agriculture.
Advancements in the field of biotechnology have increased and facilitated a more efficient and productive system for gene cloning and the production of recombinant proteins – as demonstrated by the enormous diversity of E. coli strains that are now readily available. Enhanced protein folding, greater transcriptional control and improved plasmid stability have enabled efficient gene cloning and high-throughput protein expression for structural and functional studies, in addition to industrial production.
Whether you are producing cDNA libraries, cloning methylated DNA or producing toxic proteins, there is an E. coli strain to suit your purposes. The information in this guide should help you to select the ideal strain for your experimental requirements based on their mutations and the key features they enable.
Leer más
Leer menos
References:
1. Lehman, I.R. et al. (1958) ‘Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli’, The Journal of Biological Chemistry, 233(1), pp. 163–170.
2. Crick, F.H. et al. (1961) ‘General nature of the genetic code for proteins’, Nature, 192, pp. 1227–1232. Available at: https://doi.org/10.1038/1921227a0 .
3. Jacob, F. and Monod, J. (1961) ‘Genetic regulatory mechanisms in the synthesis of proteins’, Journal of Molecular Biology, 3, pp. 318–356. Available at: https://doi.org/10.1016/s0022-2836(61)80072-7 .
4. Asefa, M. et al. (2020) ‘Escherichia coli as Model Organism and its Application in Biotechnology: A Review’, p. 10.
5. Lander, E.S. et al. (2001) ‘Initial sequencing and analysis of the human genome’, Nature, 409(6822), pp. 860–921. Available at: https://doi.org/10.1038/35057062 .
6. Riggs, A.D. (2021) ‘Making, Cloning, and the Expression of Human Insulin Genes in Bacteria: The Path to Humulin’, Endocrine Reviews, 42(3), pp. 374–380. Available at: https://doi.org/10.1210/endrev/bnaa029 .
7. Cronan, J.E. (2014) ‘Escherichia coli as an Experimental Organism’, in eLS. John Wiley & Sons, Ltd. Available at: https://doi.org/10.1002/9780470015902.a0002026.pub2 .
8. Cohen, S.N. et al. (1973) ‘Construction of biologically functional bacterial plasmids in vitro’, Proceedings of the National Academy of Sciences of the United States of America, 70(11), pp. 3240–3244. Available at: https://doi.org/10.1073/pnas.70.11.3240 .
9. Bachmann, B.J. (1972) ‘Pedigrees of some mutant strains of Escherichia coli K-12.’, Bacteriological Reviews, 36(4), pp. 525–557.
10. Daegelen, P. et al. (2009) ‘Tracing Ancestors and Relatives of Escherichia coli B, and the Derivation of B Strains REL606 and BL21(DE3)’, Journal of Molecular Biology, 394(4), pp. 634–643. Available at: https://doi.org/10.1016/j.jmb.2009.09.022 .
1. Lehman, I.R. et al. (1958) ‘Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli’, The Journal of Biological Chemistry, 233(1), pp. 163–170.
2. Crick, F.H. et al. (1961) ‘General nature of the genetic code for proteins’, Nature, 192, pp. 1227–1232. Available at: https://doi.org/10.1038/1921227a0 .
3. Jacob, F. and Monod, J. (1961) ‘Genetic regulatory mechanisms in the synthesis of proteins’, Journal of Molecular Biology, 3, pp. 318–356. Available at: https://doi.org/10.1016/s0022-2836(61)80072-7 .
4. Asefa, M. et al. (2020) ‘Escherichia coli as Model Organism and its Application in Biotechnology: A Review’, p. 10.
5. Lander, E.S. et al. (2001) ‘Initial sequencing and analysis of the human genome’, Nature, 409(6822), pp. 860–921. Available at: https://doi.org/10.1038/35057062 .
6. Riggs, A.D. (2021) ‘Making, Cloning, and the Expression of Human Insulin Genes in Bacteria: The Path to Humulin’, Endocrine Reviews, 42(3), pp. 374–380. Available at: https://doi.org/10.1210/endrev/bnaa029 .
7. Cronan, J.E. (2014) ‘Escherichia coli as an Experimental Organism’, in eLS. John Wiley & Sons, Ltd. Available at: https://doi.org/10.1002/9780470015902.a0002026.pub2 .
8. Cohen, S.N. et al. (1973) ‘Construction of biologically functional bacterial plasmids in vitro’, Proceedings of the National Academy of Sciences of the United States of America, 70(11), pp. 3240–3244. Available at: https://doi.org/10.1073/pnas.70.11.3240 .
9. Bachmann, B.J. (1972) ‘Pedigrees of some mutant strains of Escherichia coli K-12.’, Bacteriological Reviews, 36(4), pp. 525–557.
10. Daegelen, P. et al. (2009) ‘Tracing Ancestors and Relatives of Escherichia coli B, and the Derivation of B Strains REL606 and BL21(DE3)’, Journal of Molecular Biology, 394(4), pp. 634–643. Available at: https://doi.org/10.1016/j.jmb.2009.09.022 .
Leer más
Leer menos
Related links:
Leer más
Leer menos